A follower on Twitter asked me to look at two identical papers. I agreed that they looked very similar, did some searches, and found six more. All eight papers presented the same survival curves, table values, and similar line graphs. But they were published in different journals by different authors, at different institutes, on different patients, and different cancers.
In this blog post, I present to you the Mysterious Case of the Octopaper.
My investigation started with a tweet by Nirmalya Saha, who asked me to look at two identical papers, PMID: 30305431 (Liu et al., Open Biology, 2018) and PMID: 30117667 (Cai et al., J Cell Mol Med, 2018)

Of course, I was intrigued, so I compared the two papers to each other. They were from different authors at different hospitals, published in different journals, and although both groups studied miR-125a-5p expression, they did so in different types of cancer (gastric vs lung cancer).

Two papers with very similar text…
The text of the two papers, in particular the abstracts, Methods, Results, and Discussion sections were very similar to each other, as the SimTexter analysis below showed. Blocks of text in the same color show verbatim textual similarities. Only parts of the introduction and patient description were significantly different, which is not surprising because these papers described very different types of cancers.
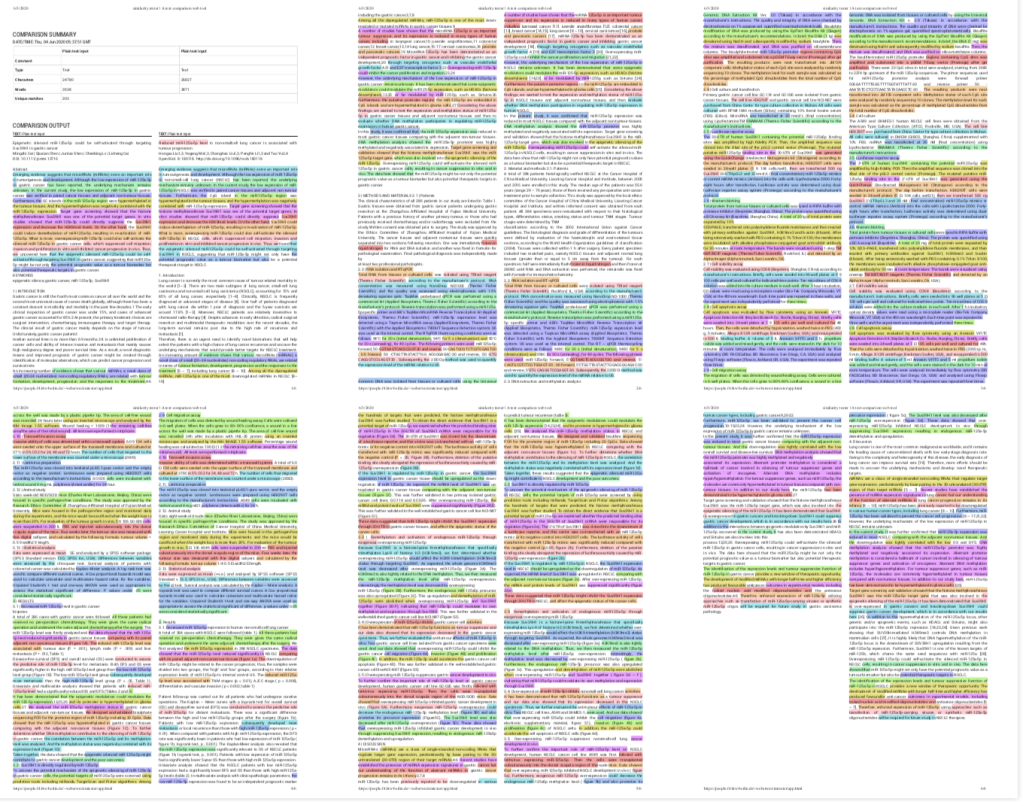
… and nearly identical figures
The figure similarities were also quite striking. Here are both Figures 1 compared to each other, with Cai et al. shown on the left, and Liu et al on the right. Even though two very different patient groups with very different types of cancer were studied, they showed a remarkably similar survival (panels B) and microRNA expression and methylation levels (panels D).
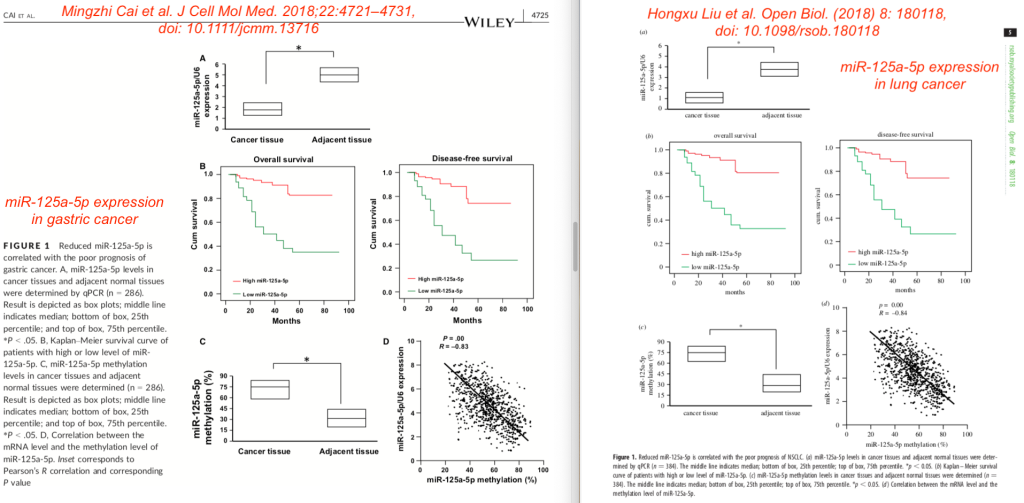
Here are both Figures 2. The layout is somewhat different, and the Cai paper has two extra photos, but what a remarkable similar set of results!

Figures 3 were also very similar, with some shuffling of the Western blots.
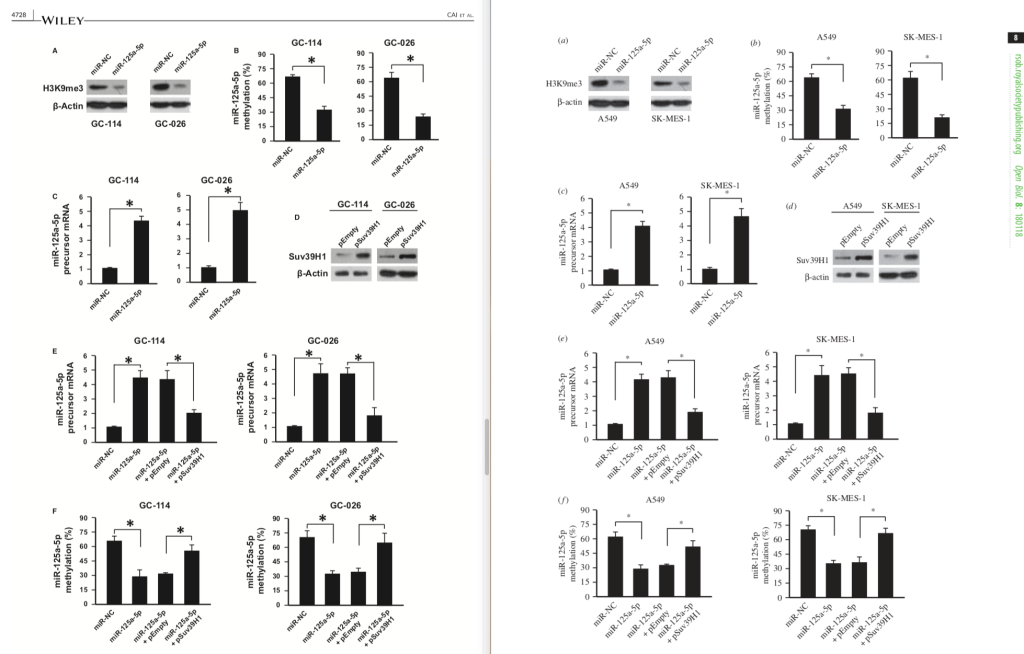
Figures 4: spot the differences.

Figures 5: again, we see the same line graphs, although now the Liu paper has been decorated with some images of mice with tumors.

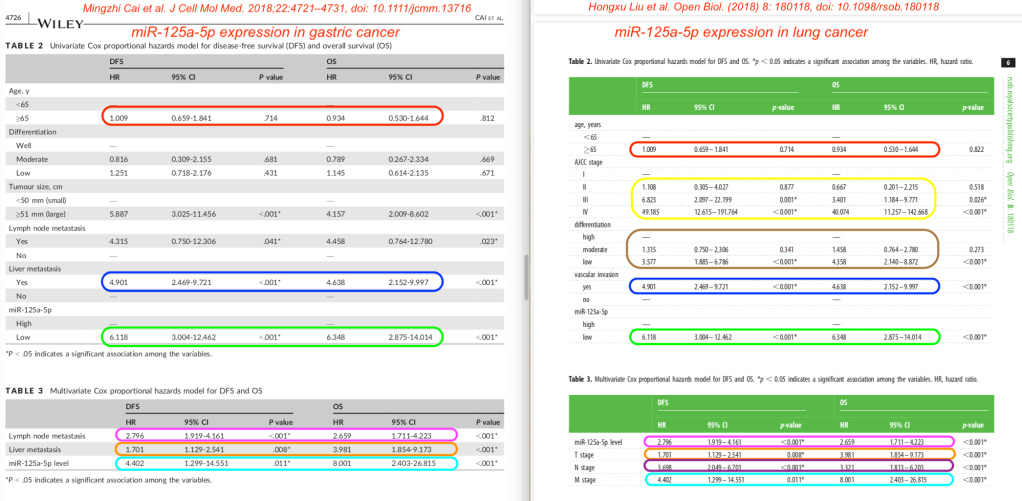
Same table values, too
Tables 2 and 3 from both papers are remarkably similar as well. The risk ratios for survival of either lung or gastric cancer are …. identical – at least if we believe these tables.
But wait – there is more
If two papers are nearly identical, there might be more. I first thought that one paper might have copied/pasted data and graphs from the other paper, but the papers were submitted pretty close to each other in time. Because of our previous work on the Tadpole Paper mill, I was starting to suspect that both papers might be the products of the same paper mill, a company that sells fabricated papers in English to doctors in China who need a paper for their promotion.
So I sailed out my data detective boat onto the big internet sea to find more papers. Based on my experience, it is hard to do successful reversed image Google searches using panels from scientific papers, so I used a different strategy. I looked for the values listed in tables 2 and 3 in Google Scholar.
This approach worked. I found six more papers that had the values that the Cai and Liu papers had in common in Tables 2 and 3. A full list of the combined eight papers is given below. All papers were published between 2017 and 2020, in five different journals, by non-overlapping authors from seven different hospitals in different cities in China, about different sets of patients with different types of cancers.
Although the figure and textual similarities were not always as striking as with the first two papers, all eight papers had very similar or identical Kaplan-Meier survival curves:
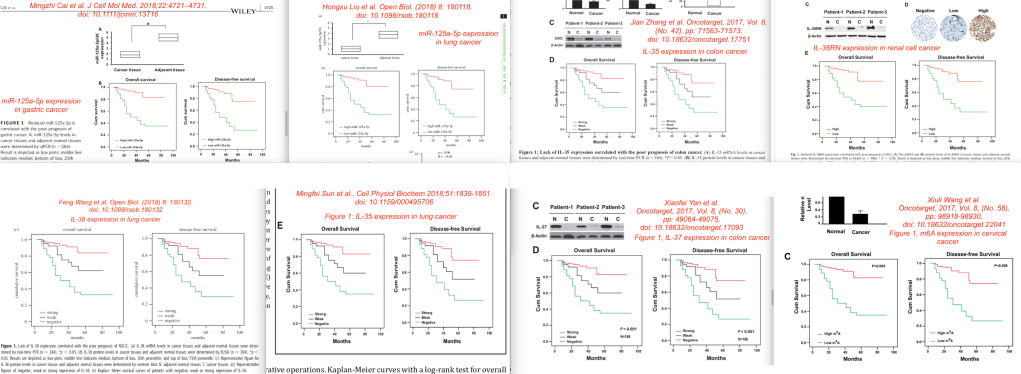
Similarly, all eight papers had identical or very similar data in tables showing the univariate and multivariate “hazards models” (risk factors for long term cancer survival).

Could this have been caught during peer review?
Although the first two papers were textually pretty similar, this would have been very hard to have been caught with a plagiarism checker. One paper was in peer review but not yet accepted or published when the second paper was submitted for peer review. The other papers are textually less similar. Most importantly, each of these papers by themselves looks legit. It took a Google Scholar search for table values to detect that all eight papers are connected.
My current suspicion is that all eight papers were generated by the same paper mill company. The company might have taken data from a real paper and adapted it to a different patient set, microRNA, or cancer type. For example, the data values all appear similar to that of an older paper, from 2015 (Zhang et al. Journal of Experimental & Clinical Cancer Research (2015), DOI 10.1186/s13046-015-0176-z), which might be the original.
In most cases, they mixed in some different figures or variations, but in the Cai and Liu papers, the company might have sold a two nearly identical copies by accident to two different group of authors.
And who knows, there might be more sets of these. Let me know below in the comments if you find another one!
List of the eight papers
| Authors | Title | Citation | DOI | Affiliation | PubPeer link |
| Mingzhi Cai, Qiuxian Chen, Juntao Shen, Chenbing Lv, Lisheng Cai | Epigenetic silenced miR-125a-5p could be self-activated through targeting Suv39H1 in gastric cancer | J Cell Mol Med. 2018;22:4721–4731 | 10.1111/jcmm.13716 | Zhangzhou Affiliated Hospital of Fujian Medical University, Zhangzhou City, China | https://pubpeer.com/publications/39A1613F4546DA16064BA441B29A0F |
| Hongxu Liu, Yegang Ma, Changhao Liu, Pengfei Li, Tao Yu | Reduced miR-125a-5p level in non-small- cell lung cancer is associated with tumour progression | Open Biol. (2018) 8: 180118 | 10.1098/rsob.180118 | Cancer Hospital of China Medical University, Shenyang, Liaoning Province, China | https://pubpeer.com/publications/5C4CE1170930C517CB660480C85DA9 |
| Xiuli Wang, Zenghui Li, Beihua Kong, Chen Song, Jianglin Cong, Jianqing Hou, Shaoguang Wang | Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer | Oncotarget, 2017, Vol. 8, (No. 58), pp: 98918-98930 | 10.18632/oncotarget.22041 | Qilu Hospital, Shandong University, Jinan, Shandong, China | https://pubpeer.com/publications/71BD811C298523B3DA169B33990572 |
| Xiaofei Yan, Jian Zhao, Rui Zhang | Interleukin-37 mediates the antitumor activity in colon cancer through β-catenin suppression | Oncotarget, 2017, Vol. 8, (No. 30), pp: 49064-49075 | 10.18632/oncotarget.17093 | Liaoning Cancer Hospital & Institute, Cancer Hospital of China Medical University, Shenyang, Liaoning Province | https://pubpeer.com/publications/BCC3ABDF2CDE9AE0DA9EED65E7E848 |
| Feng Wang, Weihua Zhang, Tianfeng Wu, Heying Chu | Reduced interleukin-38 in non-small cell lung cancer is associated with tumour progression | Open Biol. (2018) 8: 180132 | 10.1098/rsob.180132 | The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China | https://pubpeer.com/publications/EF2A21C6D3071CF820BDF72AEA23D6 |
| Jian Zhang, Tao Mao, Shuyun Wang, Dongsheng Wang, Zhaojian Niu, Zhenqing Sun, Jianli Zhang | Interleukin-35 expression is associated with colon cancer progression | Oncotarget, 2017, Vol. 8, (No. 42), pp: 71563-71573 | 10.18632/oncotarget.17751 | The Affiliated Hospital of Qingdao University, Qingdao, China | https://pubpeer.com/publications/440CE72BA5492F9C5D713FDB585913 |
| Kuangkuang Zhu, Dong Sun, Xiaoqin Zou, Ruixia Liu, Zhen Wan | Interleukin-36 receptor antagonist is associated with the progression of renal cell carcinoma | International Immunopharmacology 84 (2020) 106474 | 10.1016/j.intimp.2020.106474 | Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou, Jiangsu, China | https://pubpeer.com/publications/BAF4E0274D18F7F8DA25734CBF9053 |
| Mingfei Sun, Xianjie Zheng, Qingjiang Meng, Yanjun Dong, Guoyu Zhang, Dexin Rao, Xiaokang An, Zhongxin Yang, Lihong Pan, Shuanglin Zhang | Interleukin-35 Expression in Non-Small Cell Lung Cancer is Associated with Tumor Progression | Cell Physiol Biochem 2018;51:1839-1851 | 10.1159/000495706 | The First Affiliated Hospital of Henan University, Kaifeng City, Henan Province, China | https://pubpeer.com/publications/46F59FBF8E26449CA9F87868812E08 |

I am a Chinese and I feel ashamed when reading this. The malformed promotion rules among doctors have been there in China for a long time, and the advertisements of paper mills can be seen everywhere in hospital universities. It is really shameful for those who are making these junk papers. Thanks a lot for your job and hope it can throw a big punch to those mills.
LikeLiked by 1 person
Would it be possible to published this somewhere we could cite it? It’s really a nice piece of work that deserves citation
LikeLiked by 1 person
Well, you can cite this blog post, but it is not really material that would be accepted for publication in a scientific journal.
LikeLike
Why not? I think this should be sent as a letter to the editor. And then posted on social media of the papers involved. I’m happy to assist.
LikeLiked by 1 person
I agree with.
LikeLike
Well, it really felt ashamed of it by reading this the fabrication of the article, can anybody provide me with the criteria of plagiarism or similarity level is accepted in the publication.
LikeLike
As far as I know, no level of plagiarism should be accepted. (and I mean ZERO). I believe the issue is how to detect it efficiently.
LikeLike
That is the correct answer, but it can be confusing.
The problem is that the difference between citation, quotation and plagiarism isn’t apparent across cultural boundaries and bears better explanation than simple zero tolerance statement for plagiarism.
LikeLike
This is far worse than plagiarism! At least 7 of these 8 papers are fraudulent–fake results posing as real research.
LikeLiked by 1 person
What about the reviewers work on the manuscripts of these papers ? They also must look for plagiarism and even for autoplagiarism…..
LikeLike
I usually don’t like too much control, but this is a terrifying example of too little quality control on the editors side. Just money making from their side.
LikeLike
Editors (at least in most cases) are not paid to find plagiarism, though many of us can sense it from diction and inconsistencies. Plagiarism is squarely the fault of the writer. Lay the blame where it belongs.
LikeLike
Very sad, sad, and sad indeed.
LikeLike
This is very saddening especially in this age and time.
LikeLike
I agree with Jose Carvalho. This must be published on a scientific journal and not just restricted to
a blog. So that the editors of various journals will do a fact check as soon as the paper is submitted to them. I myselves have peer reviewed many papers and to find this kind of trick is beyond the humanly capacity of a a reviewer. The methods/algorithms you described must be translated in a software tool to unearth such an unethical practice.
LikeLike
This is not plagiarism, it’s much worse. It’s publishing fake results, a crime against all of humanity. Plagiarism does not distort scientific truth, but this type of activity could lead to decisions costing lives (if it was used as a basis for real-life decisions).
LikeLike
Very important investigation done here! Nature, Science, Cell etc. should notice this. And, its risky bussiness copiers are doing, neglecting the fact that one cancer doesen’t fit all. In the end the patient may suffer because of bad science.
LikeLiked by 1 person
I think journal should make strict provisions to have prereview check for plagiarism. It is not wise to blame reviewers. Further, there should an international active recognized platform with some legal powers to settle such cases. Otherwise such cases will be more frequent and Young Minds may be such trap.
LikeLike
Some readers of this blog are aware that I am working together with others to get retracted a fraudulent study on the breeding biology of the Basra Reed Warbler, see for backgrounds https://osf.io/5pnk7/ and https://www.researchgate.net/project/Retracting-fraudulent-articles-on-the-breeding-biology-of-the-Basra-Reed-Warbler-Acrocephalus-griseldis
The Basra Reed Warbler is an Endangered bird species which is almost exclusively breeding in Iraq, see http://datazone.birdlife.org/species/factsheet/basra-reed-warbler-acrocephalus-griseldis The fraudulent study is the first one on major aspects of its breeding biology. The study is fraudulent because the raw research data, collected in the field in Iraq, do not exist. The first author is willing to retract the study.
It has turned out that it is extremely difficult to get published in a peer-reviewed journal an article which is mainly based on the findings of two reports about this case (both reports can be downloaded from
https://osf.io/5pnk7/ ). The manuscript has in the meanwhile been submitted to in total 26 different peer-reviewed journals, both in the field of ornithology / ecology / nature conservation and in the field of research ethics. Only 2 journals, both in the field of research ethics, were willing to send it our for peer-review, all other submissions were desk-rejected. Several EiC’s refused to communicate about any of the findings in both reports and/or used invalid motives for their desk rejection.
Other desk-rejections contained very valuable comments. Franz Bairlein, EiC of the Journal of Ornithology https://www.springer.com/journal/10336 stated on 21 December 2018: ‘We agree with you that the Basra Reed-Warbler paper by Al-Sheikhly et al. is a case of scientific misconduct’. Javier Seoane, EiC of Ardeola https://www.ardeola.org/en/ stated on 17 March 2020: ‘The manuscript is a compelling report of a worrying case of scientific misconduct’. I am at the moment processing the extensive comments of 2 anonymous reviewers of journal #26. I will submit a new version to journal #27.
It is thus understandable that Elisabeth Bik states in her comment: “it is not really material that would be accepted for publication in a scientific journal.”
LikeLike
Shameful act against humanity, just for ordinary promotion to be reckoned with and not solving problems at all. I am happy that this type of blog is exposing a lot of shady deals about publication and promotion in academics. We still have a lot to learn from this. I can’t imagine this. Very sad.
LikeLike
I noticed that all papers except one (published in “International Immunopharmacology” in 2020) were published in the period 2017-18. The fact that there is a new published version of this paper after a “silent” year could imply that the paper mill is producing a new round of falsified versions of this paper. I wouldn’t be surprised if a few more version have been submitted for review but not published yet.
I don’t know if it would be possible (and how) to raise awareness about this problem. Maybe a published article, highlighting the main similarities between these papers could help other scientist and reviewers to identify more cases.
Also, I assume that after the initial “success” of their business, the paper-mill most probably has produced other fake papers with multiple versions of each. It is extremely alarming.
LikeLike
Is any media (not private blogs) that take care about your finding?
LikeLike
Is your finding published in media other then blogs?
LikeLike
Hi. I really commend the great work. This has inspired me to crawl pubmed systematically [automatically] to identify clusters of fraudulent papers. I can’t believe this is a one-off and it would be nice to catch more of this. Thanks.
LikeLike
This is very discouraging to all the hard-working young scientists out there. Would strongly suggest you submit this to a major general journal – write to Editors and I’m sure you get good response.
LikeLike